@ WFS,World Fossil Society,Riffin T Sajeev,Russel T Sajeev
Scientists at Scripps Research and their collaborators have created microorganisms that may recapitulate key features of organisms thought to have lived billions of years ago, allowing them to explore questions about how life evolved from inanimate molecules to single-celled organisms to the complex, multicellular lifeforms we see today.
By studying one of these engineered organisms-a bacterium whose genome consists of both ribonucleic acid (RNA) and deoxyribonucleic acid (DNA)-the scientists hope to shed light on the early evolution of genetic material, including the theorized transition from a world where most life relied solely on the genetic molecule RNA to one where DNA serves as the primary storehouse of genetic information.
Using a second engineered organism, a genetically modified yeast containing an endosymbiotic bacterium, they hope to better understand the origins of cellular power plants called mitochondria. Mitochondria provide essential energy for the cells of eukaryotes, a broad group of organisms-including humans-that possesses complex, nucleus-containing cells.
A genetically modified yeast containing an endosymbiotic bacterium.Credit: Scripps Research
The researchers report engineering the microbes in two papers, one published October 29, 2018 in the Proceedings of the National Academy of Sciences (PNAS) and another published August 30, 2018 in Journal of the American Chemical Society (JACS).
“These engineered organisms will allow us to probe two key theories about major milestones in the evolution of living organisms-the transition from the RNA world to the DNA world and the transition from prokaryotes to eukaryotes with mitochondria,” says Peter Schultz, PhD, senior author on the papers and president of Scripps Research. “Access to readily manipulated laboratory models enables us to seek answers to questions about early evolution that were previously intractable.”
The origins of life on Earth have been a human fascination for millennia. Scientists have traced the arc of life back several billion years and concluded that the simplest forms of life emerged from Earth’s primordial chemical soup and subsequently evolved over the eons into organisms of greater and greater complexity. A monumental leap came with the emergence of DNA, a molecule that stores all of the information required to replicate life and directs cellular machinery to do its bidding primarily by generating RNA, which in turn directs the synthesis of proteins, the molecular workhorses in cells.
In the 1960s, Carl Woese and Leslie Orgel, along with DNA pioneer Francis Crick, proposed that before DNA, organisms relied on RNA to carry genetic information, a molecule similar to but far less stable than DNA, that can also catalyze chemical reactions like proteins. “In science class, students learn that DNA leads to RNA which in turn leads to proteins-that’s a central dogma of biology-but the RNA world hypothesis turns that on its head,” says Angad Mehta, PhD, first author of the new papers and a postdoctoral research associate at Scripps Research. “For the RNA world hypothesis to be true, you have to somehow get from RNA to a DNA genome, yet how that might have happened is still a very big question among scientists.”
One possibility is that the transition proceeded through a kind of microbial missing link, a replicating organism that stored genetic information as RNA. For the JACS study, the Scripps Research-led team created Escherichia coli bacteria that partially build their DNA with ribonucleotides, the molecular building blocks typically used to build RNA. These engineered genomes contained up to 50 percent RNA, thus simultaneously representing a new type of synthetic organism and possibly a throwback to billions of years ago.
Mehta cautions that their work so far has focused on characterizing this chimeric RNA-DNA genome and its effect on bacterial growth and replication but hasn’t explicitly explored questions about the transition from the RNA world to the DNA world. But, he says, the fact that E. coli with half its genome comprised of RNA can survive and replicate is remarkable and seems to support the possibility of the existence of evolutionarily transitional organisms possessing hybrid RNA-DNA genomes. The Scripps Research team is now studying how the mixed genomes of their engineered E. coli function and plans to use the bacteria to explore a number of evolutionary questions.
For instance, one question is whether the presence of RNA leads to rapid genetic drift-large changes in gene sequence in a population over time. Scientists theorize that massive genetic drift occurred quickly during early evolution, and the presence in the genome of RNA could help explain how genetic change occurred so quickly.
In the paper published in PNAS, the researchers report engineering another laboratory model for an evolutionary milestone thought to have occurred more than 1.5 billion years ago. They created a yeast dependent for energy on bacteria living inside it as a beneficial parasite or “endosymbiont.” This composite organism will allow them to investigate the ancient origins of mitochondria-tiny, bacteria-like organelles that produce chemical energy within the cells of all higher organisms.
Mitochondria are widely thought to have evolved from ordinary bacteria that were captured by larger, single-celled organisms. They carry out several key functions in cells. Most importantly, they serve as oxygen reactors, using O2 to make cells’ basic unit of chemical energy, the molecule ATP. As crucial as mitochondria are to cells, their origins remain somewhat mysterious, although there are clear hints of descent from a more independent organism, widely assumed to have been a bacterium.
Mitochondria have a double-membrane structure like that of some bacteria, and-again, like bacteria-contain their own DNA. Analyses of the mitochondrial genome suggest that it shares an ancient ancestor with modern Rickettsia bacteria, which can live within the cells of their hosts and cause disease. Stronger support for the bacterial origin of mitochondria theory would come from experiments showing that independent bacteria could indeed be transformed, in an evolution-like progression, into mitochondria-like symbionts. To that end, the Scripps Research scientists engineered E. coli bacteria that could live in, depend upon, and provide key assistance to, cells of Saccharomyces cerevisiae, also known as baker’s yeast.
The researchers started by modifying E. coli to lack the gene encoding thiamin, making the bacteria dependent on the yeast cells for this essential vitamin. At the same time, they added to the bacteria a gene for ADP/ATP translocase, a transporter protein, so that ATP produced within the bacterial cells would be supplied to their yeast-cell hosts-mimicking the central function of real mitochondria. The team also modified the yeast so that their own mitochondria were deficient at supplying ATP. Thus the yeast would be dependent on the bacteria for normal, mitochondria-based ATP production.
The team found that some of the engineered bacteria, after being modified with surface proteins to protect them from being destroyed in the yeast, lived and proliferated in harmony with their hosts for more than 40 generations and appeared to be viable indefinitely. “The modified bacteria seem to accumulate new mutations within the yeast to better adapt to their new surroundings,” says Schultz.
With this system established, the team will try to evolve the E. colito become mitochondria-like organelles. For the new E. coliendosymbiont, adapting to life inside yeast could allow it an opportunity to radically slim its genome. A typical E. coli bacterium, for example, has several thousand genes, whereas mitochondria have evolved a stripped-down set of just 37.
The Scripps Research team rounded out the study with further gene-subtraction experiments, and the results were promising: they found they could eliminate not just the E. coli thiamin gene but also the genes underlying the production of the metabolic molecule NAD and the amino acid serine, and still get a viable symbiosis.
“We are now well on our way to showing that we can delete the genes for making all 20 amino acids, which comprise a significant part of the E. coli genome,” says Schultz. “Once we’ve achieved that, we’ll move on to deleting genes for the syntheses of cofactors and nucleotides, and within a few years we hope to be able to get a truly minimal endosymbiotic genome.”
The researchers also hope to use similar endosymbiont-host systems to investigate other important episodes in evolution, such as the origin of chloroplasts, light-absorbing organelles that have a mitochondria-like role in supplying energy to plants.
Journal References:Angad P. Mehta, Yiyang Wang, Sean A. Reed, Lubica Supekova, Tsotne Javahishvili, John C. Chaput, Peter G. Schultz. Bacterial Genome Containing Chimeric DNA–RNA Sequences. Journal of the American Chemical Society, 2018; 140 (36): 11464 DOI: 10.1021/jacs.8b07046
- Angad P. Mehta, Lubica Supekova, Jian-Hua Chen, Kersi Pestonjamasp, Paul Webster, Yeonjin Ko, Scott C. Henderson, Gerry McDermott, Frantisek Supek, Peter G. Schultz. Engineering yeast endosymbionts as a step toward the evolution of mitochondria. Proceedings of the National Academy of Sciences, 2018; 201813143 DOI: 10.1073/pnas.1813143115
@ WFS,World Fossil Society,Riffin T Sajeev,Russel T Sajeev



 November 1st, 2018
November 1st, 2018  Riffin
Riffin 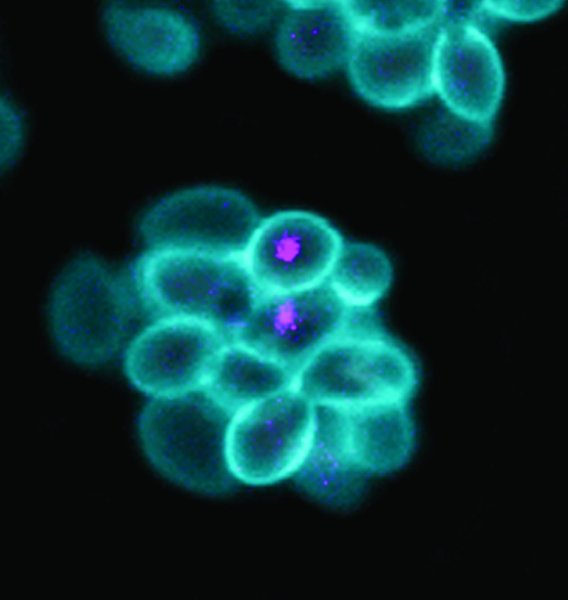
 Posted in
Posted in  Tags:
Tags: 